class: center, middle, inverse, title-slide # OpenML: Connecting R to the Machine Learning Platform OpenML ## useR! 2017 tutorial - <a href="http://bit.ly/2tcb2b7" class="uri">http://bit.ly/2tcb2b7</a> ### Joaquin Vanschoren, Bernd Bischl, Heidi Seibold ### <em>If you haven’t done so yet, create an account on OpenML.org, and install the OpenML R package and either packages farff or RWeka</em> --- <!-- For this to work, install xaringan (devtools::install_github('yihui/xaringan')) --> ## Preliminaries - If you haven't done so yet, create an account on [OpenML.org](www.openml.org). - If you haven't done so yet, install the OpenML R package and one of the packages farff or RWeka: ```r install.packages("OpenML") install.packages("farff") # or install.packages("RWeka") ``` ```r library("OpenML") ``` - If something is not clear / you have a question / you have a problem, please **let us know**! - We will have lots of practicals, if you are faster than others, you can check out https://www.openml.org/guide or help others. --- Help  --- ## OpenML useR! Tutorial Learning goals: - Understand the **potentials** of OpenML - Use the OpenML **online platform** and the **R package** + Creating, uploading and downloading + Running algorithms on OpenML tasks - Know about cool OpenML **projects** and how to **get involved** --- ### Installation and configuration <!-- [3 minutes, Joaquin] --> You need OpenML and an ARFF reader ```r install.packages(c("OpenML","farff")) ``` ```r library("OpenML") ``` --- You also need an OpenML API key to talk to the server ```r setOMLConfig(apikey = "c1994bdb7ecb3c6f3c8f3b35f4b47f1f") ``` - Find your own key in your OpenML profile  --- Permanently save your API disk to your config file (~/.openml/config) ```r saveOMLConfig(apikey = "c1994...47f1f", overwrite=TRUE) ``` Other configuration options: - `server`: default https://www.openml.org/api/v1 - `cachedir`: cache directory - `verbosity`: 0 (normal) - 2 (debug) - `arff.reader`: 'farff' (default) or 'RWeka' - `confirm.upload`: default FALSE View your configuration ```r getOMLConfig() ``` ``` ## OpenML configuration: ## server : http://www.openml.org/api/v1 ## cachedir : /home/bischl/.openml/cache ## verbosity : 1 ## arff.reader : farff ## confirm.upload : FALSE ## apikey : ***************************af579 ``` --- class:inverse ### *Practical* <!-- [5 minutes, Joaquin] --> - Install the OpenML R package (if you haven't done so yet) - Add your API-key to your config file - Configure OpenML to your liking --- ### Listing datasets <!-- [7 minutes, Joaquin] --> ```r datasets = listOMLDataSets() # get first 5K results (LIMIT option) colnames(datasets) ``` ``` ## [1] "data.id" ## [2] "name" ## [3] "version" ## [4] "status" ## [5] "format" ## [6] "tags" ## [7] "majority.class.size" ## [8] "max.nominal.att.distinct.values" ## [9] "minority.class.size" ## [10] "number.of.classes" ## [11] "number.of.features" ## [12] "number.of.instances" ## [13] "number.of.instances.with.missing.values" ## [14] "number.of.missing.values" ## [15] "number.of.numeric.features" ## [16] "number.of.symbolic.features" ``` --- ### Listing datasets ```r datasets[1:15, c(1,2,11,12)] # subset for compact display ``` ``` ## data.id name number.of.features number.of.instances ## 1 1 anneal 39 898 ## 2 2 anneal 39 898 ## 3 3 kr-vs-kp 37 3196 ## 4 4 labor 17 57 ## 5 5 arrhythmia 280 452 ## 6 6 letter 17 20000 ## 7 7 audiology 70 226 ## 8 8 liver-disorders 7 345 ## 9 9 autos 26 205 ## 10 10 lymph 19 148 ## 11 11 balance-scale 5 625 ## 12 12 mfeat-factors 217 2000 ## 13 13 breast-cancer 10 286 ## 14 14 mfeat-fourier 77 2000 ## 15 15 breast-w 10 699 ``` --- ### Listing tasks ```r tasks = listOMLTasks() # limits results again colnames(tasks)[1:20] ``` ``` ## [1] "task.id" "task.type" ## [3] "data.id" "name" ## [5] "status" "format" ## [7] "estimation.procedure" "evaluation.measures" ## [9] "target.feature" "cost.matrix" ## [11] "source.data.labeled" "target.feature.event" ## [13] "target.feature.left" "target.feature.right" ## [15] "quality.measure" "tags" ## [17] "majority.class.size" "max.nominal.att.distinct.values" ## [19] "minority.class.size" "number.of.classes" ``` --- ### Listing tasks ```r tasks[1:5, 1:9] ``` ``` ## task.id task.type data.id name status format ## 1 1 Supervised Classification 1 anneal active ARFF ## 2 2 Supervised Classification 2 anneal active ARFF ## 3 3 Supervised Classification 3 kr-vs-kp active ARFF ## 4 4 Supervised Classification 4 labor active ARFF ## 5 5 Supervised Classification 5 arrhythmia active ARFF ## estimation.procedure evaluation.measures target.feature ## 1 10-fold Crossvalidation predictive_accuracy class ## 2 10-fold Crossvalidation predictive_accuracy class ## 3 10-fold Crossvalidation predictive_accuracy class ## 4 10-fold Crossvalidation predictive_accuracy class ## 5 10-fold Crossvalidation predictive_accuracy class ``` --- ### Listing tasks ```r listOMLTasks(data.name = "GesturePhaseSegmentationRAW")[,1:8] ``` ``` ## task.id task.type data.id name ## 1 128503 Clustering 4537 GesturePhaseSegmentationRAW ## 2 145676 Supervised Classification 4537 GesturePhaseSegmentationRAW ## 3 146567 Supervised Classification 4537 GesturePhaseSegmentationRAW ## status format estimation.procedure evaluation.measures ## 1 active ARFF 50 times Clustering <NA> ## 2 active ARFF 10-fold Crossvalidation area_under_roc_curve ## 3 active ARFF 10-fold Crossvalidation predictive_accuracy ``` --- ### Listing flows ```r flows = listOMLFlows() flows[56:63, c(1,2,6,7)] ``` ``` ## flow.id full.name uploader tags ## 56 56 weka.ZeroR(1) 1 study_7 ## 57 57 weka.OneR(1) 1 ## 58 58 weka.NaiveBayes(1) 1 ## 59 59 weka.JRip(1) 1 ## 60 60 weka.J48(1) 1 ## 61 61 weka.REPTree(1) 1 ## 62 62 weka.DecisionStump(1) 1 ## 63 63 weka.HoeffdingTree(1) 1 ``` --- ### Listing runs and evaluations Runs and evaluations need at least one of: task.id, flow.id, run.id, uploader.id, tag ```r runs = listOMLRuns(task.id = 146567L) head(runs) ``` ``` ## run.id task.id setup.id flow.id uploader error.message tags ## 1 3893285 146567 1930168 6770 2 <NA> useR17 ## 2 3893865 146567 442005 6748 2 <NA> useR17 ## 3 3902928 146567 24109 4840 2 <NA> useR17 ## 4 3930588 146567 24109 4840 27 <NA> useR17 ## 5 3963816 146567 24109 4840 27 <NA> useR17 ## 6 3963925 146567 24109 4840 27 <NA> useR17 ``` --- ### Listing runs and evaluations ```r evals = listOMLRunEvaluations(task.id = 146567L) evals[, c("flow.name", "predictive.accuracy")] ``` ``` ## flow.name predictive.accuracy ## 1 mlr.classif.randomForest(30) 0.944551 ## 2 mlr.classif.rpart(37) 0.577719 ## 3 mlr.classif.rpart(24) 0.577719 ## 4 mlr.classif.rpart(24) 0.577719 ## 5 mlr.classif.rpart(37) 0.577719 ## 6 mlr.classif.rpart(24) 0.577719 ``` --- class:inverse ### *Practical* <!-- [5 minutes, Joaquin] --> - List all datasets having between 100K and 200K observations and <= 5 features - List all regression tasks corresponding to data sets having between 50 and 55 observations. - Find the data.id for dataset 'GesturePhaseSegmentationRAW' - List all `predictive.accuracy` evaluations for task 146567 --- class:inverse ### *Solution* - List all datasets having between 100K and 200K observations and <= 5 features ```r datasets = listOMLDataSets(number.of.instances = c(100000, 200000), number.of.features = c(1, 5)) str(datasets, list.len = 10) ``` ``` ## 'data.frame': 1 obs. of 16 variables: ## $ data.id : int 1509 ## $ name : chr "walking-activity" ## $ version : int 1 ## $ status : chr "active" ## $ format : chr "ARFF" ## $ tags : chr "" ## $ majority.class.size : int 21991 ## $ max.nominal.att.distinct.values : int 22 ## $ minority.class.size : int 911 ## $ number.of.classes : int 22 ## [list output truncated] ``` --- class:inverse ### *Solution* - List all regression tasks corresponding to data sets having between 50 and 55 observations. ```r tasks = listOMLTasks(task.type = "Supervised Regression", number.of.instances = c(50, 55)) tasks[1:10, 1:5] ``` ``` ## task.id task.type data.id name status ## 1 2283 Supervised Regression 192 vineyard active ## 2 2316 Supervised Regression 228 elusage active ## 3 4979 Supervised Regression 660 rabe_265 active ## 4 4985 Supervised Regression 668 witmer_census_1980 active ## 5 5002 Supervised Regression 691 chscase_vine1 active ## 6 5003 Supervised Regression 692 rabe_131 active ## 7 5015 Supervised Regression 707 sleuth_case1201 active ## 8 5032 Supervised Regression 1090 MercuryinBass active ## 9 5037 Supervised Regression 1096 FacultySalaries active ## 10 5045 Supervised Regression 192 vineyard active ``` --- class:inverse ### *Solution* - Find the data.id for dataset 'GesturePhaseSegmentationRAW' ```r ds = listOMLDataSets(data.name = "GesturePhaseSegmentationRAW") str(ds, list.len = 10) ``` ``` ## 'data.frame': 1 obs. of 15 variables: ## $ data.id : int 4537 ## $ name : chr "GesturePhaseSegmentationRAW" ## $ version : int 1 ## $ status : chr "active" ## $ format : chr "ARFF" ## $ tags : chr "" ## $ majority.class.size : int 2950 ## $ minority.class.size : int 1 ## $ number.of.classes : int 6 ## $ number.of.features : int 20 ## [list output truncated] ``` --- class:inverse ### *Solution* - List all `predictive.accuracy` evaluations for task 146567 ```r evals = listOMLRunEvaluations(task.id = 146567) evals[, c(1:5, 11:14)] ``` ``` ## run.id task.id setup.id flow.id flow.name ## 1 3893285 146567 1930168 6770 mlr.classif.randomForest(30) ## 2 3893865 146567 442005 6748 mlr.classif.rpart(37) ## 3 3963816 146567 24109 4840 mlr.classif.rpart(24) ## 4 3963925 146567 24109 4840 mlr.classif.rpart(24) ## 5 3965307 146567 442005 6748 mlr.classif.rpart(37) ## 6 3978948 146567 24109 4840 mlr.classif.rpart(24) ## area.under.roc.curve average.cost f.measure kappa ## 1 0.963933 0 0.944283 0.927491 ## 2 0.715748 0 0.525545 0.427405 ## 3 0.715748 0 0.525545 0.427405 ## 4 0.715748 0 0.525545 0.427405 ## 5 0.715748 0 0.525545 0.427405 ## 6 0.715748 0 0.525545 0.427405 ``` --- ### Downloading datasets <!-- [10 minutes, Joaquin] --> ```r gesture.data = getOMLDataSet(data.id = 4537L) gesture.data$desc ``` ``` ## ## Data Set "GesturePhaseSegmentationRAW" :: (Version = 1, OpenML ID = 4537) ## Creator(s) : Renata Cristina Barros Madeo (Madeo; R. C. B.) Priscilla Koch Wagner (Wagner; P. K.) Sarajane Marques Peres (Peres; S. M.) {renata.si; priscilla.wagner; sarajane} at usp.br http://each.uspnet.usp.br/sarajane/ ## Default Target Attribute: phase ``` ```r gesture.data$data[1:5, c(1:5,20)] ``` ``` ## lhx lhy lhz rhx rhy phase ## 0 5.347435 4.363681 1.501913 5.258967 4.319263 Rest ## 1 4.869622 4.254210 1.556133 5.240113 4.346338 Rest ## 2 5.357447 4.364039 1.500969 5.238928 4.347924 Rest ## 3 4.942886 4.281878 1.546513 5.111436 4.229660 Rest ## 4 5.003160 4.278530 1.542866 4.985812 4.182155 Rest ``` --- ### Downloading datasets For convenience, you can also download by name ```r getOMLDataSet(data.name = "iris") ``` ``` ## ## Data Set "iris" :: (Version = 3, OpenML ID = 969) ## Default Target Attribute: binaryClass ``` --- ### Downloading tasks ```r task = getOMLTask(task.id = 146567L) task ``` ``` ## ## OpenML Task 146567 :: (Data ID = 4537) ## Task Type : Supervised Classification ## Data Set : GesturePhaseSegmentationRAW :: (Version = 1, OpenML ID = 4537) ## Target Feature(s) : phase ## Estimation Procedure : Stratified crossvalidation (1 x 10 folds) ## Evaluation Measure(s): predictive_accuracy ``` --- ### Downloading flows ```r flow = getOMLFlow(flow.id = 100L) flow ``` ``` ## ## Flow "weka.J48" :: (Version = 2, Flow ID = 100) ## External Version : Weka_3.7.5_9117 ## Dependencies : Weka_3.7.5 ## Number of Flow Parameters: 12 ## Number of Flow Components: 0 ``` --- ### Downloading runs ```r run = getOMLRun(run.id = 3893285L) run ``` ``` ## ## OpenML Run 3893285 :: (Task ID = 146567, Flow ID = 6770) ## User ID : 2 ## Tags : useR17 ## Learner : mlr.classif.randomForest(30) ## Task type: Supervised Classification ``` --- ### Caching - The package caches most objects on disk (cachedir in config) - Results of listing calls are cached in memory - You can also pre-fill the cache with objects, especially useful on clusters ```r populateOMLCache(data.ids = 1:2, task.ids = 11:12) ``` ``` cache/ ├── datasets │ └── 37 │ ├── dataset.arff │ └── description.xml ├── flows │ └── 100 │ └── flow.xml ├── runs │ └── 1815885 │ ├── predictions.arff │ └── run.xml └── tasks └─── 146567 ├── datasplits.arff └── task.xml ``` --- class:inverse ### *Practical* <!-- [5 minutes, Joaquin] --> - Download task with task Id 59 - Extract the dataset from this task Hint: read ?OMLTask and look for "input" --- class:inverse ### *Practical* - Download task with task Id 59 ```r task = getOMLTask(task.id = 59) task ``` ``` ## ## OpenML Task 59 :: (Data ID = 61) ## Task Type : Supervised Classification ## Data Set : iris :: (Version = 1, OpenML ID = 61) ## Target Feature(s) : class ## Tags : basic, study_1, study_41, study_50, study_7, under100k, under1m ## Estimation Procedure : Stratified crossvalidation (1 x 10 folds) ## Evaluation Measure(s): predictive_accuracy ``` --- class:inverse ### *Practical* - Extract the dataset from this task ```r task$input$data ``` ``` ## ## Data Set "iris" :: (Version = 1, OpenML ID = 61) ## Collection Date : 1936 ## Creator(s) : R.A. Fisher ## Default Target Attribute: class ``` ```r head(task$input$data$data) ``` ``` ## sepallength sepalwidth petallength petalwidth class ## 0 5.1 3.5 1.4 0.2 Iris-setosa ## 1 4.9 3.0 1.4 0.2 Iris-setosa ## 2 4.7 3.2 1.3 0.2 Iris-setosa ## 3 4.6 3.1 1.5 0.2 Iris-setosa ## 4 5.0 3.6 1.4 0.2 Iris-setosa ## 5 5.4 3.9 1.7 0.4 Iris-setosa ``` --- class: inverse <!-- background-image: url(https://c1.staticflickr.com/5/4049/4468213356_07ffffd287_z.jpg) --> background-image: url(slides_tutorial_files/cat.jpg) background-size: cover # BREAK TIME ??? Image credit: [Dave Dugdale](https://flic.kr/p/7NQLaj) --- ## Intro to mlr <!-- [15 minutes, Bernd] --> mlr = General umbrella package for ML in R with standardized interface <img src="slides_tutorial_files/mlr.jpg" width="800px" /> - Project home page: https://github.com/mlr-org/mlr - 8-10 main developers, quite a few contributors - Extensive online tutorial available, look there first - Can ask questions in the github issue tracker Install for the following examples: ```r install.packages(c("randomForest", "kernlab")) ``` `mlr` is atomatically installed when installing `OpenML` --- ## Intro to mlr - Classification, regression, survival, clustering, cost-sensitive, multilabel - Includes > 160 basic learning algorithms - Unified interface for the basic building blocks: tasks, learners, resampling, hyperparameters - Reflections: nearly all objects are queryable, i.e. you can ask for their properties and program on them - Programmed in an OO fashion in S3 (everything is an object) - Makes extensions and generic algorithms easy <img src="slides_tutorial_files/ml_abstraction-crop.png" width="500px" /> --- ### mlr - Train, predict, performance ```r task = makeClassifTask(data = iris, target = "Species") print(task) ``` ``` ## Supervised task: iris ## Type: classif ## Target: Species ## Observations: 150 ## Features: ## numerics factors ordered ## 4 0 0 ## Missings: FALSE ## Has weights: FALSE ## Has blocking: FALSE ## Classes: 3 ## setosa versicolor virginica ## 50 50 50 ## Positive class: NA ``` --- ### mlr - Train, predict, performance ```r lrn = makeLearner("classif.rpart", minsplit = 5) lrn ``` ``` ## Learner classif.rpart from package rpart ## Type: classif ## Name: Decision Tree; Short name: rpart ## Class: classif.rpart ## Properties: twoclass,multiclass,missings,numerics,factors,ordered,prob,weights,featimp ## Predict-Type: response ## Hyperparameters: xval=0,minsplit=5 ``` ```r model = train(lrn, task, subset = seq(1, 150, by = 2)) model ``` ``` ## Model for learner.id=classif.rpart; learner.class=classif.rpart ## Trained on: task.id = iris; obs = 75; features = 4 ## Hyperparameters: xval=0,minsplit=5 ``` --- ### mlr - Train, predict, performance ```r pred = predict(model, task, subset = seq(2, 150, by = 2)) pred ``` ``` ## Prediction: 75 observations ## predict.type: response ## threshold: ## time: 0.01 ## id truth response ## 2 2 setosa setosa ## 4 4 setosa setosa ## 6 6 setosa setosa ## 8 8 setosa setosa ## 10 10 setosa setosa ## 12 12 setosa setosa ## ... (75 rows, 3 cols) ``` ```r perf = performance(pred, measures = list(mmce, ber)) perf ``` ``` ## mmce ber ## 0.05333333 0.05333333 ``` --- ### mlr - Resample - Crossvalidation, subsampling, bootstrapping, etc., with a single command - Get a container object with + Mean performances and performances per resampling iteration + Predictions + Models (if you want that) ```r lrn = makeLearner("classif.rpart", minsplit = 5) rdesc = makeResampleDesc("CV", iters = 2) # or use "cv2" object r = resample(lrn, task, rdesc, measures = list(mmce, ber), models = TRUE) r ``` ``` ## Resample Result ## Task: iris ## Learner: classif.rpart ## Aggr perf: mmce.test.mean=0.04,ber.test.mean=0.0412 ## Runtime: 0.0162368 ``` --- ### mlr - Resample ```r r$aggr ``` ``` ## mmce.test.mean ber.test.mean ## 0.04000000 0.04121965 ``` ```r r$measures.test ``` ``` ## iter mmce ber ## 1 1 0.02666667 0.02380952 ## 2 2 0.05333333 0.05862978 ``` --- ### mlr - Resample ```r head(as.data.frame(r$pred)) ``` ``` ## id truth response iter set ## 1 3 setosa setosa 1 test ## 2 6 setosa setosa 1 test ## 3 7 setosa setosa 1 test ## 4 9 setosa setosa 1 test ## 5 10 setosa setosa 1 test ## 6 17 setosa setosa 1 test ``` ```r r$models ``` ``` ## [[1]] ## Model for learner.id=classif.rpart; learner.class=classif.rpart ## Trained on: task.id = iris; obs = 75; features = 4 ## Hyperparameters: xval=0,minsplit=5 ## ## [[2]] ## Model for learner.id=classif.rpart; learner.class=classif.rpart ## Trained on: task.id = iris; obs = 75; features = 4 ## Hyperparameters: xval=0,minsplit=5 ``` --- ### mlr - Benchmarking and Model Comparison - Run one command to compare multiple learners on multiple data sets - Get a (mergeable) container object with + Mean performances and performances per resampling iteration + Predictions + Models (if you want that) ```r # these are predefined in mlr for toying around: tasks = list(iris.task, sonar.task) learners = makeLearners(c("classif.rpart", "classif.randomForest")) br = benchmark(learners, tasks, cv2) br # again, you can access container in various ways ``` ``` ## task.id learner.id mmce.test.mean ## 1 iris-example classif.rpart 0.06000000 ## 2 iris-example classif.randomForest 0.04666667 ## 3 Sonar-example classif.rpart 0.27403846 ## 4 Sonar-example classif.randomForest 0.16826923 ``` --- ### mlr - More features and outlook Uncovered features - Many different NA imputation techniques - Many feature filters - Imbalancy correction (e.g SMOTE) - Use wrappers to extend learner functionality - Simple nested resampling - Efficient tuning: Bayesian optimization with mlrMBO and iterated F-racing with irace - Multi-criteria optimization - Feature selection through wrappers (forward, backward) - Ensembles, generic bagging and stacking What will come next - Anomaly detection / one-class Classification - Functional data handling - Time series forecasting - Efficient pipelining --- ### mlr GSOC teaser: Pipelines / composable preproc ops - You cannot run this with the current release! - Will be finished in Oct 2017 (after GSOC) ```r pipeline1 = cpoImputeMedian() %>>% cpoDropConstanst() %>>% cpoPca() %>>% cpoFilterGainRatio() pipeline2 = pipeline1 %>>% makeLearner("classif.rpart") crossval(pipeline1, task) ``` - Pipelines can be tuned as normal learners! - Multiplexing and feature unions are possible! - Have a look here if you are curious: https://github.com/mlr-org/mlr/pull/1827 --- class:center,middle # Back to OpenML --- ### Running and uploading <!-- [10 minutes, Bernd] --> Create a run: 1. Define a learner using the `mlr` package 2. Apply to a task using `runTaskMlr()` ```r # create a randomForest learner lrn = makeLearner("classif.randomForest", mtry = 2) # download a task (or get from cache) task = getOMLTask(task.id = 37) # runs the learner locally, uses benchmark internally run.mlr = runTaskMlr(task, lrn) ``` - The `run.mlr` object contains three slots + `run`: contains the information of the run, i.e., the hyperparameter values and the learner predictions. + `bmr`: the `BenchmarkResult` object containing the results of the learner that is applied on the task. + `flow`: contains information about the algorithm. --- ### Running and uploading ```r run.mlr ``` ``` ## $run ## ## OpenML Run NA :: (Task ID = 37, Flow ID = NA) ## ## $bmr ## task.id learner.id acc.test.join timetrain.test.sum ## 1 diabetes classif.randomForest 0.7708333 2.939 ## timepredict.test.sum ## 1 0.146 ## ## $flow ## ## Flow "mlr.classif.randomForest" :: (Version = NA, Flow ID = NA) ## External Version : R_3.3.2-v2.b88a2294 ## Dependencies : R_3.3.2, OpenML_1.4, mlr_2.11, randomForest_4.6.12 ## Number of Flow Parameters: 20 ## Number of Flow Components: 0 ## ## attr(,"class") ## [1] "OMLMlrRun" ``` <!-- From a didactical standpoint I would not include this: --> <!-- - Extract the `BenchmarkResult` object via ```r convertOMLMlrRunToBMR(run.mlr) ``` ``` ## task.id learner.id acc.test.join timetrain.test.sum ## 1 diabetes classif.randomForest 0.7708333 2.939 ## timepredict.test.sum ## 1 0.146 ``` --> --- ### Running and uploading Upload run to OpenML server: ```r run.id = uploadOMLRun(run.mlr) ``` ```r run.id ``` ``` ## [1] 3828647 ``` - The server assigns a `run.id` which can be used to - download the run: `getOMLRun(run.id)`, or - look up the run online on https://www.openml.org/r/3828647. - Server auto-computes many evaluation measures from predictions <!-- - It is also possible to upload runs with specific tags using the `tags` argument, so that finding the run with a specific tag becomes easier. --> --- ### Running and uploading Let's check that we can get our run back: ```r getOMLRun(run.id) ``` ``` ## ## OpenML Run 3828647 :: (Task ID = 37, Flow ID = 6770) ## User ID : 970 ## Tags : useR17 ## Learner : mlr.classif.randomForest(30) ## Task type: Supervised Classification ``` --- ### OpenML <-> mlr converters - `convertOMLDataSetToMlr` Convert an OpenML data set to mlr task. - `convertMlrTaskToOMLDataSet` Converts a mlr task to an OpenML data set. - `convertOMLFlowToMlr` Converts a flow to a mlr learner. - `convertOMLMlrRunToBMR` Convert 'OMLMlrRun's to a 'BenchmarkResult'. - `convertOMLRunToBMR` Convert an OpenML run set to a benchmark result for mlr. - `convertOMLTaskToMlr` Convert an OpenML task to mlr. --- class:inverse ### *Practical* <!-- [25 minutes, Bernd] --> - Run your favorite learner/algorithm (from mlr) on task 146567 that you already downloaded. + You can run `listLearners()` to find appropriate learners + Or go to mlr's appendix in the web tutorial to see a table - Upload your run to OpenML. Add the tag "useR17". Hint: ```r run.id = uploadOMLRun(myrun, tag = "useR17") ``` - Check if the upload worked by going to the website. Check if the tag was added (you can still add it on the website if you forgot during the upload). - Check the predictive performance of the run (it may take a while before the server has calculated the performance measures `\(\rightarrow\)` give it some time). For fast solvers: - Run a 2nd learner and compare them on the webpage - Add tuning to a learner with an mlr TuneWrapper (much harder, look at the tutorial for this!) --- class:inverse ### *Solution* - Run your favorite learner/algorithm (from mlr) on task 146567 that you already downloaded. ```r # list mlr algos: subset for compact display listLearners(warn.missing.packages=FALSE)[10:14, c(1,2,4,6)] ``` ``` ## class name package type ## 10 classif.ksvm Support Vector Machines kernlab classif ## 11 classif.lda Linear Discriminant Analysis MASS classif ## 12 classif.logreg Logistic Regression stats classif ## 13 classif.lssvm Least Squares Support Vector Machine kernlab classif ## 14 classif.lvq1 Learning Vector Quantization class classif ``` ```r # run favorite learner lrn = makeLearner("classif.rpart", minsplit = 5) myrun = runTaskMlr(task, lrn) ``` - Upload your run to OpenML. Add the tag "useR17". ```r myrun.id = uploadOMLRun(myrun, tags = "useR17") ``` --- class:inverse ### *Solution* - Check if the upload worked by going to the website. Also check if the tag was added (you can also still add it on the website). Use the value from `myrun.id` and go to https://www.openml.org/r/3829199. - Check the predictive performance of the run by looking at several evaluation measures. - Scroll down to *Evaluation measures*. --- class:inverse ### *Solution* You can also get the results via ```r str(listOMLRunEvaluations(run.id = myrun.id), list.len = 15) ``` ``` ## 'data.frame': 1 obs. of 30 variables: ## $ run.id : int 3829199 ## $ task.id : int 37 ## $ setup.id : int 442005 ## $ flow.id : int 6748 ## $ flow.name : chr "mlr.classif.rpart(37)" ## $ flow.version : chr "37" ## $ flow.source : chr "mlr" ## $ learner.name : chr "classif.rpart" ## $ data.name : chr "diabetes" ## $ upload.time : chr "2017-07-03 18:44:05" ## $ area.under.roc.curve : num 0.698 ## $ average.cost : num 0 ## $ f.measure : num 0.734 ## $ kappa : num 0.407 ## $ kb.relative.information.score: num 308 ## [list output truncated] ``` --- ## Tags <!-- [10 minutes, Heidi] --> Use tags to sort and find data, tasks, flows and runs.  ```r uploadOMLRun(myrun, tags = c("tag1", "tag2")) ``` --- ## Studies Studies are an extension of tags and get their own website. Tag must be `study_XX`  ??? - With tasks we can e.g. combine several runs and find them again and make a little benchmarking study. - The information what the tag means probably not obvious to other OpenML users: this is why we created studies - Study = tag + website with study information --- ## Studies  ```r uploadOMLRun(myrun, tags = "study_30") ``` --- ## Evaluations ```r evals = listOMLRunEvaluations(tag = "study_30") evals[1:3, c("data.name", "learner.name", "predictive.accuracy")] ``` ``` ## data.name learner.name predictive.accuracy ## 1 diabetes classif.randomForest 0.772135 ## 2 sonar classif.randomForest 0.817308 ## 3 haberman classif.randomForest 0.748366 ``` 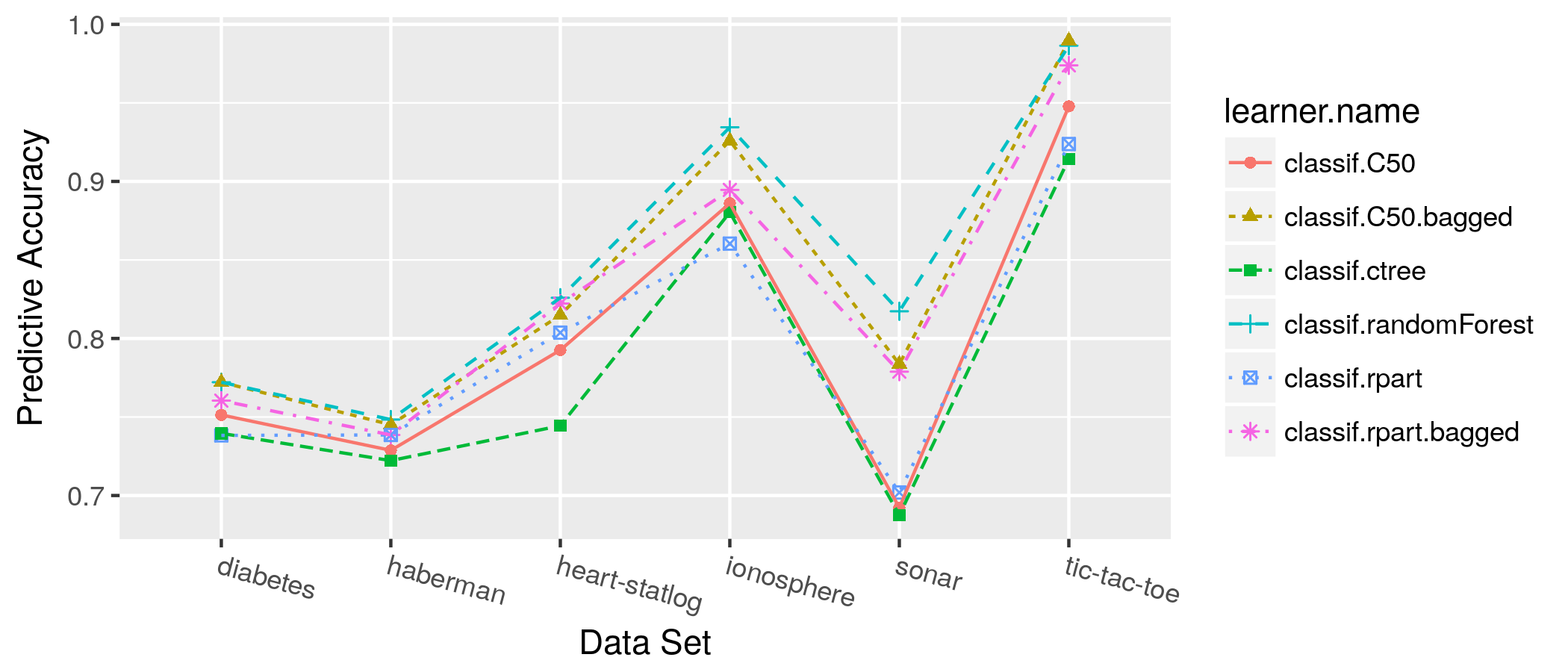<!-- --> --- class:inverse ### *Practical* <!-- [10 minutes, Heidi] --> - List the names of all data sets in study_27 - Summarize the performance results of study_27 (look at predictive accuracy) > Predictive accuracy is the percentage of instances that are classified correctly. > (Information like this can be found on [openml.org/a](https://www.openml.org/a)) - Bonus questions for fast solvers: + What are the two different versions of ksvm? + What do the different setup.id s mean? --- class:inverse ### *Solution* <!-- [10 minutes, Heidi] --> - List the names of all data sets in study_27 - Summarize the performance results of study_27 ```r *evals = listOMLRunEvaluations(tag = "study_27") evals$setup.id = as.factor(evals$setup.id) library("ggplot2") ggplot(evals, aes(x = setup.id, y = predictive.accuracy, color = data.name, group = data.name)) + geom_point() + geom_line() + facet_grid(~ flow.name, scales = "free") ``` 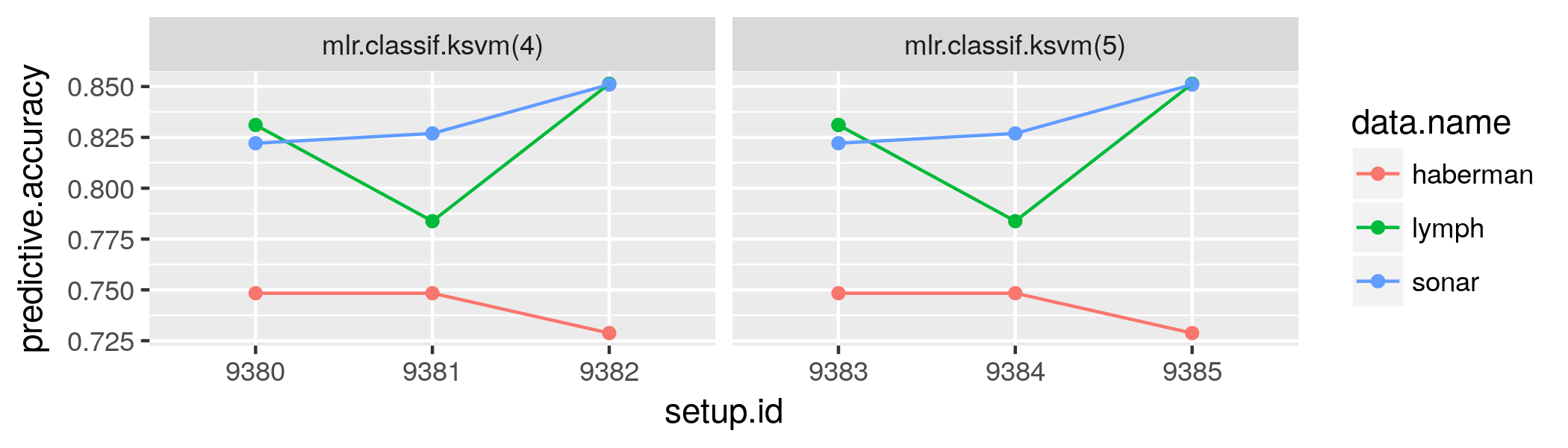<!-- --> --- class:inverse Why are there two different versions of the flow? ```r fids = unique(evals$flow.id) flws = lapply(fids, getOMLFlow) flws ``` ``` ## [[1]] ## ## Flow "mlr.classif.ksvm" :: (Version = 4, Flow ID = 4704) ## External Version : R_3.3.1-v2.bf0ac616 ## Dependencies : R_3.3.1, OpenML_1.0, mlr_2.9, kernlab_0.9.24 ## Number of Flow Parameters: 19 ## Number of Flow Components: 0 ## ## [[2]] ## ## Flow "mlr.classif.ksvm" :: (Version = 5, Flow ID = 4705) ## External Version : R_3.2.2-v2.f878bda4 ## Dependencies : R_3.2.2, OpenML_1.0, mlr_2.10, kernlab_0.9.24 ## Number of Flow Parameters: 19 ## Number of Flow Components: 0 ``` --- class:inverse What are the different setup IDs? ```r rids = evals$run.id runs = lapply(rids, getOMLRun) params = lapply(runs, getOMLRunParList) params[[1]] ``` ``` ## This is a 'OMLRunParList' with the following parameters: ## name value component ## 1: fit FALSE NA ``` ```r params[[5]] ``` ``` ## This is a 'OMLRunParList' with the following parameters: ## name value component ## 1: type kbb-svc NA ## 2: fit FALSE NA ``` ```r params[[19]] ``` ``` ## This is a 'OMLRunParList' with the following parameters: ## name value component ## 1: type spoc-svc NA ## 2: fit FALSE NA ``` The difference seems to be the type. --- class:inverse Let's add the type info to the evals data frame. ```r evals$type = sapply(params, function(x) ifelse(is.null(x$type$value), NA, x$type$value)) evals$type ``` ``` ## [1] NA NA NA "kbb-svc" "kbb-svc" "kbb-svc" ## [7] "spoc-svc" "spoc-svc" "spoc-svc" NA NA NA ## [13] NA "kbb-svc" "kbb-svc" "kbb-svc" "spoc-svc" "spoc-svc" ## [19] "spoc-svc" ``` ```r evals$type[is.na(evals$type)] = getDefaults( getParamSet( * convertOMLFlowToMlr(flws[[1]]) ) )$type evals$type ``` ``` ## [1] "C-svc" "C-svc" "C-svc" "kbb-svc" "kbb-svc" "kbb-svc" ## [7] "spoc-svc" "spoc-svc" "spoc-svc" "C-svc" "C-svc" "C-svc" ## [13] "C-svc" "kbb-svc" "kbb-svc" "kbb-svc" "spoc-svc" "spoc-svc" ## [19] "spoc-svc" ``` --- class:inverse Now we can make a more understandable plot. ```r ggplot(evals, aes(x = type, y = predictive.accuracy, color = data.name, group = data.name)) + geom_point() + geom_line() + facet_grid(~ flow.name, scales = "free") ``` <!-- --> --- background-image: url(slides_tutorial_files/cool_stuff_text.png) background-size: 70% auto ## Cool stuff people are already doing with OpenML <!-- [15 minutes, Heidi] --> ??? Image-credit: https://commons.wikimedia.org --- ### OpenML Bot - Currently completing 100.000+ runs per day on Azure - Exploring hyperparameters of xgboost, ranger, and other popular machine learning algorithms - Using 75 datasets from study_14  --- ### OpenML meta learning: Making defaults great again! - Choose between different performance measures (AUC, RMSE, ...) - Predict the pareto front for this measure and the training time - E. g. for xgboost: Prediction for hyperparameters on a new dataset, which will outperform the defaults  --- ### OpenML in drug discovery Predict which drugs will inhibit certain proteins (and hence viruses, parasites, ...)  <!-- <img src="slides_tutorial_files/qsar.pdf" width="4200" height="4200"> --> <!-- <object data="slides_tutorial_files/qsar.pdf" type="application/pdf" width="700px" height="700px"> --> <!-- <embed src="slides_tutorial_files/qsar.pdf"> --> <!-- This browser does not support PDFs. Please download the PDF to view it: <a href="slides_tutorial_files/qsar.pdf">Download PDF</a>.</p> --> <!-- </embed> --> <!-- </object> --> --- class: inverse <!-- background-image: url(https://c1.staticflickr.com/6/5477/14101220086_a633ec9674_c.jpg) --> background-image: url(slides_tutorial_files/fish.jpg) background-size: cover # Contributors needed! https://github.com/openml/OpenML/wiki/How-to-contribute <!-- [10 minutes, Heidi] --> ??? Image credit: [Papahānaumokuākea Marine National Monument](https://flic.kr/p/neCvxt) --- class: inverse, center, middle  Thanks to all the great folks who have been contributing to OpenML and the R package.