Note
Go to the end to download the full example code.
Measuring runtimes for Scikit-learn models¶
The runtime of machine learning models on specific datasets can be a deciding factor on the choice of algorithms, especially for benchmarking and comparison purposes. OpenML’s scikit-learn extension provides runtime data from runs of model fit and prediction on tasks or datasets, for both the CPU-clock as well as the actual wallclock-time incurred. The objective of this example is to illustrate how to retrieve such timing measures, and also offer some potential means of usage and interpretation of the same.
It should be noted that there are multiple levels at which parallelism can occur.
At the outermost level, OpenML tasks contain fixed data splits, on which the defined model/flow is executed. Thus, a model can be fit on each OpenML dataset fold in parallel using the n_jobs parameter to run_model_on_task or run_flow_on_task (illustrated under Case 2 & 3 below).
The model/flow specified can also include scikit-learn models that perform their own parallelization. For instance, by specifying n_jobs in a Random Forest model definition (covered under Case 2 below).
The sklearn model can further be an HPO estimator and contain it’s own parallelization. If the base estimator used also supports parallelization, then there’s at least a 2-level nested definition for parallelization possible (covered under Case 3 below).
We shall cover these 5 representative scenarios for:
(Case 1) Retrieving runtimes for Random Forest training and prediction on each of the cross-validation folds
(Case 2) Testing the above setting in a parallel setup and monitor the difference using runtimes retrieved
(Case 3) Comparing RandomSearchCV and GridSearchCV on the above task based on runtimes
(Case 4) Running models that don’t run in parallel or models which scikit-learn doesn’t parallelize
(Case 5) Running models that do not release the Python Global Interpreter Lock (GIL)
# License: BSD 3-Clause
import openml
import numpy as np
from matplotlib import pyplot as plt
from joblib.parallel import parallel_backend
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV
Preparing tasks and scikit-learn models¶
task_id = 167119
task = openml.tasks.get_task(task_id)
print(task)
# Viewing associated data
n_repeats, n_folds, n_samples = task.get_split_dimensions()
print(
"Task {}: number of repeats: {}, number of folds: {}, number of samples {}.".format(
task_id,
n_repeats,
n_folds,
n_samples,
)
)
# Creating utility function
def print_compare_runtimes(measures):
for repeat, val1 in measures["usercpu_time_millis_training"].items():
for fold, val2 in val1.items():
print(
"Repeat #{}-Fold #{}: CPU-{:.3f} vs Wall-{:.3f}".format(
repeat, fold, val2, measures["wall_clock_time_millis_training"][repeat][fold]
)
)
OpenML Classification Task
==========================
Task Type Description: https://www.openml.org/tt/TaskType.SUPERVISED_CLASSIFICATION
Task ID..............: 167119
Task URL.............: https://www.openml.org/t/167119
Estimation Procedure.: crossvalidation
Target Feature.......: class
# of Classes.........: 3
Cost Matrix..........: Available
Task 167119: number of repeats: 1, number of folds: 10, number of samples 1.
Case 1: Running a Random Forest model on an OpenML task¶
We’ll run a Random Forest model and obtain an OpenML run object. We can see the evaluations recorded per fold for the dataset and the information available for this run.
clf = RandomForestClassifier(n_estimators=10)
run1 = openml.runs.run_model_on_task(
model=clf,
task=task,
upload_flow=False,
avoid_duplicate_runs=False,
)
measures = run1.fold_evaluations
print("The timing and performance metrics available: ")
for key in measures.keys():
print(key)
print()
print(
"The performance metric is recorded under `predictive_accuracy` per "
"fold and can be retrieved as: "
)
for repeat, val1 in measures["predictive_accuracy"].items():
for fold, val2 in val1.items():
print("Repeat #{}-Fold #{}: {:.4f}".format(repeat, fold, val2))
print()
0%| | 0.00/16.1k [00:00<?, ?B/s]
100%|██████████| 16.1k/16.1k [00:00<00:00, 104kB/s]
100%|██████████| 16.1k/16.1k [00:00<00:00, 104kB/s]
The timing and performance metrics available:
usercpu_time_millis_training
wall_clock_time_millis_training
usercpu_time_millis_testing
usercpu_time_millis
wall_clock_time_millis_testing
wall_clock_time_millis
predictive_accuracy
The performance metric is recorded under `predictive_accuracy` per fold and can be retrieved as:
Repeat #0-Fold #0: 0.7767
Repeat #0-Fold #1: 0.7657
Repeat #0-Fold #2: 0.7764
Repeat #0-Fold #3: 0.7834
Repeat #0-Fold #4: 0.7818
Repeat #0-Fold #5: 0.7760
Repeat #0-Fold #6: 0.7700
Repeat #0-Fold #7: 0.7724
Repeat #0-Fold #8: 0.7802
Repeat #0-Fold #9: 0.7692
The remaining entries recorded in measures are the runtime records related as:
usercpu_time_millis = usercpu_time_millis_training + usercpu_time_millis_testing
wall_clock_time_millis = wall_clock_time_millis_training + wall_clock_time_millis_testing
The timing measures recorded as *_millis_training contain the per repeat-per fold timing incurred for the execution of the .fit() procedure of the model. For usercpu_time_* the time recorded using time.process_time() is converted to milliseconds and stored. Similarly, time.time() is used to record the time entry for wall_clock_time_*. The *_millis_testing entry follows the same procedure but for time taken for the .predict() procedure.
# Comparing the CPU and wall-clock training times of the Random Forest model
print_compare_runtimes(measures)
Repeat #0-Fold #0: CPU-247.832 vs Wall-247.864
Repeat #0-Fold #1: CPU-252.822 vs Wall-252.825
Repeat #0-Fold #2: CPU-250.680 vs Wall-250.682
Repeat #0-Fold #3: CPU-245.216 vs Wall-245.126
Repeat #0-Fold #4: CPU-242.658 vs Wall-242.698
Repeat #0-Fold #5: CPU-244.409 vs Wall-244.412
Repeat #0-Fold #6: CPU-254.070 vs Wall-254.085
Repeat #0-Fold #7: CPU-247.591 vs Wall-246.531
Repeat #0-Fold #8: CPU-242.659 vs Wall-242.568
Repeat #0-Fold #9: CPU-244.182 vs Wall-244.197
Case 2: Running Scikit-learn model on an OpenML task in parallel¶
Redefining the model to allow parallelism with n_jobs=2 (2 cores)
clf = RandomForestClassifier(n_estimators=10, n_jobs=2)
run2 = openml.runs.run_model_on_task(
model=clf, task=task, upload_flow=False, avoid_duplicate_runs=False
)
measures = run2.fold_evaluations
# The wall-clock time recorded per fold should be lesser than Case 1 above
print_compare_runtimes(measures)
Repeat #0-Fold #0: CPU-273.833 vs Wall-167.753
Repeat #0-Fold #1: CPU-267.301 vs Wall-155.562
Repeat #0-Fold #2: CPU-253.158 vs Wall-153.932
Repeat #0-Fold #3: CPU-258.289 vs Wall-153.039
Repeat #0-Fold #4: CPU-250.039 vs Wall-144.498
Repeat #0-Fold #5: CPU-258.718 vs Wall-155.345
Repeat #0-Fold #6: CPU-268.424 vs Wall-155.095
Repeat #0-Fold #7: CPU-252.020 vs Wall-143.993
Repeat #0-Fold #8: CPU-251.033 vs Wall-154.636
Repeat #0-Fold #9: CPU-264.367 vs Wall-155.200
Running a Random Forest model on an OpenML task in parallel (all cores available):
# Redefining the model to use all available cores with `n_jobs=-1`
clf = RandomForestClassifier(n_estimators=10, n_jobs=-1)
run3 = openml.runs.run_model_on_task(
model=clf, task=task, upload_flow=False, avoid_duplicate_runs=False
)
measures = run3.fold_evaluations
# The wall-clock time recorded per fold should be lesser than the case above,
# if more than 2 CPU cores are available. The speed-up is more pronounced for
# larger datasets.
print_compare_runtimes(measures)
Repeat #0-Fold #0: CPU-322.441 vs Wall-132.588
Repeat #0-Fold #1: CPU-320.288 vs Wall-130.413
Repeat #0-Fold #2: CPU-313.292 vs Wall-121.843
Repeat #0-Fold #3: CPU-313.107 vs Wall-124.611
Repeat #0-Fold #4: CPU-318.973 vs Wall-130.680
Repeat #0-Fold #5: CPU-312.666 vs Wall-123.321
Repeat #0-Fold #6: CPU-317.181 vs Wall-131.123
Repeat #0-Fold #7: CPU-308.808 vs Wall-124.559
Repeat #0-Fold #8: CPU-322.069 vs Wall-122.303
Repeat #0-Fold #9: CPU-308.425 vs Wall-126.721
We can now observe that the ratio of CPU time to wallclock time is lower than in case 1. This happens because joblib by default spawns subprocesses for the workloads for which CPU time cannot be tracked. Therefore, interpreting the reported CPU and wallclock time requires knowledge of the parallelization applied at runtime.
Running the same task with a different parallel backend. Joblib provides multiple backends: {loky (default), multiprocessing, dask, threading, sequential}. The backend can be explicitly set using a joblib context manager. The behaviour of the job distribution can change and therefore the scale of runtimes recorded too.
with parallel_backend(backend="multiprocessing", n_jobs=-1):
run3_ = openml.runs.run_model_on_task(
model=clf, task=task, upload_flow=False, avoid_duplicate_runs=False
)
measures = run3_.fold_evaluations
print_compare_runtimes(measures)
Repeat #0-Fold #0: CPU-372.033 vs Wall-422.385
Repeat #0-Fold #1: CPU-374.382 vs Wall-342.099
Repeat #0-Fold #2: CPU-372.110 vs Wall-427.754
Repeat #0-Fold #3: CPU-374.666 vs Wall-419.478
Repeat #0-Fold #4: CPU-352.880 vs Wall-307.285
Repeat #0-Fold #5: CPU-378.178 vs Wall-314.064
Repeat #0-Fold #6: CPU-367.198 vs Wall-354.472
Repeat #0-Fold #7: CPU-365.928 vs Wall-371.067
Repeat #0-Fold #8: CPU-356.117 vs Wall-217.919
Repeat #0-Fold #9: CPU-341.743 vs Wall-206.166
The CPU time interpretation becomes ambiguous when jobs are distributed over an unknown number of cores or when subprocesses are spawned for which the CPU time cannot be tracked, as in the examples above. It is impossible for OpenML-Python to capture the availability of the number of cores/threads, their eventual utilisation and whether workloads are executed in subprocesses, for various cases that can arise as demonstrated in the rest of the example. Therefore, the final interpretation of the runtimes is left to the user.
Case 3: Running and benchmarking HPO algorithms with their runtimes¶
We shall now optimize a similar RandomForest model for the same task using scikit-learn’s HPO support by using GridSearchCV to optimize our earlier RandomForest model’s hyperparameter n_estimators. Scikit-learn also provides a refit_time_ for such HPO models, i.e., the time incurred by training and evaluating the model on the best found parameter setting. This is included in the wall_clock_time_millis_training measure recorded.
from sklearn.model_selection import GridSearchCV
clf = RandomForestClassifier(n_estimators=10, n_jobs=2)
# GridSearchCV model
n_iter = 5
grid_pipe = GridSearchCV(
estimator=clf,
param_grid={"n_estimators": np.linspace(start=1, stop=50, num=n_iter).astype(int).tolist()},
cv=2,
n_jobs=2,
)
run4 = openml.runs.run_model_on_task(
model=grid_pipe, task=task, upload_flow=False, avoid_duplicate_runs=False, n_jobs=2
)
measures = run4.fold_evaluations
print_compare_runtimes(measures)
Repeat #0-Fold #0: CPU-6600.493 vs Wall-3643.300
Repeat #0-Fold #1: CPU-6174.082 vs Wall-3426.261
Repeat #0-Fold #2: CPU-6480.912 vs Wall-3651.825
Repeat #0-Fold #3: CPU-6569.833 vs Wall-3707.602
Repeat #0-Fold #4: CPU-6096.366 vs Wall-3396.607
Repeat #0-Fold #5: CPU-6098.743 vs Wall-3448.627
Repeat #0-Fold #6: CPU-6156.255 vs Wall-3450.285
Repeat #0-Fold #7: CPU-6052.819 vs Wall-3349.450
Repeat #0-Fold #8: CPU-6107.637 vs Wall-3481.000
Repeat #0-Fold #9: CPU-6459.292 vs Wall-3673.058
Like any optimisation problem, scikit-learn’s HPO estimators also generate a sequence of configurations which are evaluated, using which the best found configuration is tracked throughout the trace. The OpenML run object stores these traces as OpenMLRunTrace objects accessible using keys of the pattern (repeat, fold, iterations). Here fold implies the outer-cross validation fold as obtained from the task data splits in OpenML. GridSearchCV here performs grid search over the inner-cross validation folds as parameterized by the cv parameter. Since GridSearchCV in this example performs a 2-fold cross validation, the runtime recorded per repeat-per fold in the run object is for the entire fit() procedure of GridSearchCV thus subsuming the runtimes of the 2-fold (inner) CV search performed.
# We earlier extracted the number of repeats and folds for this task:
print("# repeats: {}\n# folds: {}".format(n_repeats, n_folds))
# To extract the training runtime of the first repeat, first fold:
print(run4.fold_evaluations["wall_clock_time_millis_training"][0][0])
# repeats: 1
# folds: 10
3643.3000564575195
To extract the training runtime of the 1-st repeat, 4-th (outer) fold and also to fetch the parameters and performance of the evaluations made during the 1-st repeat, 4-th fold evaluation by the Grid Search model.
_repeat = 0
_fold = 3
print(
"Total runtime for repeat {}'s fold {}: {:4f} ms".format(
_repeat, _fold, run4.fold_evaluations["wall_clock_time_millis_training"][_repeat][_fold]
)
)
for i in range(n_iter):
key = (_repeat, _fold, i)
r = run4.trace.trace_iterations[key]
print(
"n_estimators: {:>2} - score: {:.3f}".format(
r.parameters["parameter_n_estimators"], r.evaluation
)
)
Total runtime for repeat 0's fold 3: 3707.601786 ms
n_estimators: 1 - score: 0.764
n_estimators: 13 - score: 0.799
n_estimators: 25 - score: 0.800
n_estimators: 37 - score: 0.803
n_estimators: 50 - score: 0.803
Scikit-learn’s HPO estimators also come with an argument refit=True as a default. In our previous model definition it was set to True by default, which meant that the best found hyperparameter configuration was used to refit or retrain the model without any inner cross validation. This extra refit time measure is provided by the scikit-learn model as the attribute refit_time_. This time is included in the wall_clock_time_millis_training measure.
For non-HPO estimators, wall_clock_time_millis = wall_clock_time_millis_training + wall_clock_time_millis_testing.
For HPO estimators, wall_clock_time_millis = wall_clock_time_millis_training + wall_clock_time_millis_testing + refit_time.
This refit time can therefore be explicitly extracted in this manner:
def extract_refit_time(run, repeat, fold):
refit_time = (
run.fold_evaluations["wall_clock_time_millis"][repeat][fold]
- run.fold_evaluations["wall_clock_time_millis_training"][repeat][fold]
- run.fold_evaluations["wall_clock_time_millis_testing"][repeat][fold]
)
return refit_time
for repeat in range(n_repeats):
for fold in range(n_folds):
print(
"Repeat #{}-Fold #{}: {:.4f}".format(
repeat, fold, extract_refit_time(run4, repeat, fold)
)
)
Repeat #0-Fold #0: 793.9892
Repeat #0-Fold #1: 594.5659
Repeat #0-Fold #2: 795.6455
Repeat #0-Fold #3: 844.9187
Repeat #0-Fold #4: 608.1171
Repeat #0-Fold #5: 568.8848
Repeat #0-Fold #6: 616.5302
Repeat #0-Fold #7: 588.4402
Repeat #0-Fold #8: 597.1401
Repeat #0-Fold #9: 733.7892
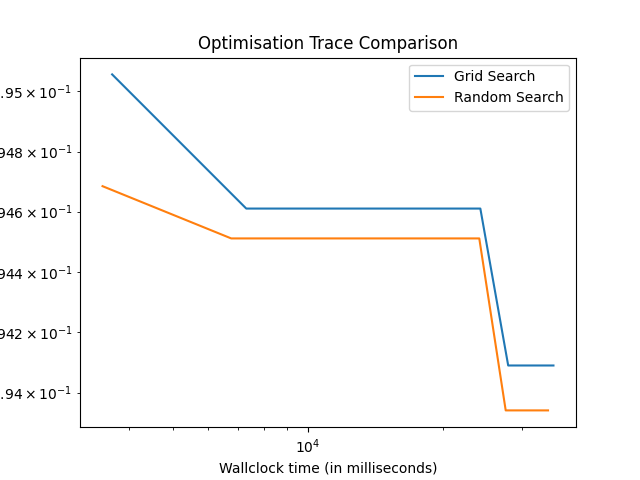
Along with the GridSearchCV already used above, we demonstrate how such optimisation traces can be retrieved by showing an application of these traces - comparing the speed of finding the best configuration using RandomizedSearchCV and GridSearchCV available with scikit-learn.
# RandomizedSearchCV model
rs_pipe = RandomizedSearchCV(
estimator=clf,
param_distributions={
"n_estimators": np.linspace(start=1, stop=50, num=15).astype(int).tolist()
},
cv=2,
n_iter=n_iter,
n_jobs=2,
)
run5 = openml.runs.run_model_on_task(
model=rs_pipe, task=task, upload_flow=False, avoid_duplicate_runs=False, n_jobs=2
)
Since for the call to openml.runs.run_model_on_task the parameter
n_jobs is set to its default None, the evaluations across the OpenML folds
are not parallelized. Hence, the time recorded is agnostic to the n_jobs
being set at both the HPO estimator GridSearchCV as well as the base
estimator RandomForestClassifier in this case. The OpenML extension only records the
time taken for the completion of the complete fit() call, per-repeat per-fold.
This notion can be used to extract and plot the best found performance per
fold by the HPO model and the corresponding time taken for search across
that fold. Moreover, since n_jobs=None for openml.runs.run_model_on_task
the runtimes per fold can be cumulatively added to plot the trace against time.
def extract_trace_data(run, n_repeats, n_folds, n_iter, key=None):
key = "wall_clock_time_millis_training" if key is None else key
data = {"score": [], "runtime": []}
for i_r in range(n_repeats):
for i_f in range(n_folds):
data["runtime"].append(run.fold_evaluations[key][i_r][i_f])
for i_i in range(n_iter):
r = run.trace.trace_iterations[(i_r, i_f, i_i)]
if r.selected:
data["score"].append(r.evaluation)
break
return data
def get_incumbent_trace(trace):
best_score = 1
inc_trace = []
for i, r in enumerate(trace):
if i == 0 or (1 - r) < best_score:
best_score = 1 - r
inc_trace.append(best_score)
return inc_trace
grid_data = extract_trace_data(run4, n_repeats, n_folds, n_iter)
rs_data = extract_trace_data(run5, n_repeats, n_folds, n_iter)
plt.clf()
plt.plot(
np.cumsum(grid_data["runtime"]), get_incumbent_trace(grid_data["score"]), label="Grid Search"
)
plt.plot(
np.cumsum(rs_data["runtime"]), get_incumbent_trace(rs_data["score"]), label="Random Search"
)
plt.xscale("log")
plt.yscale("log")
plt.xlabel("Wallclock time (in milliseconds)")
plt.ylabel("1 - Accuracy")
plt.title("Optimisation Trace Comparison")
plt.legend()
plt.show()
Case 4: Running models that scikit-learn doesn’t parallelize¶
Both scikit-learn and OpenML depend on parallelism implemented through joblib. However, there can be cases where either models cannot be parallelized or don’t depend on joblib for its parallelism. 2 such cases are illustrated below.
Running a Decision Tree model that doesn’t support parallelism implicitly, but using OpenML to parallelize evaluations for the outer-cross validation folds.
dt = DecisionTreeClassifier()
run6 = openml.runs.run_model_on_task(
model=dt, task=task, upload_flow=False, avoid_duplicate_runs=False, n_jobs=2
)
measures = run6.fold_evaluations
print_compare_runtimes(measures)
Repeat #0-Fold #0: CPU-83.224 vs Wall-83.225
Repeat #0-Fold #1: CPU-87.205 vs Wall-87.216
Repeat #0-Fold #2: CPU-85.151 vs Wall-85.152
Repeat #0-Fold #3: CPU-86.270 vs Wall-86.271
Repeat #0-Fold #4: CPU-82.451 vs Wall-82.462
Repeat #0-Fold #5: CPU-81.686 vs Wall-81.688
Repeat #0-Fold #6: CPU-85.637 vs Wall-85.657
Repeat #0-Fold #7: CPU-87.602 vs Wall-87.603
Repeat #0-Fold #8: CPU-86.538 vs Wall-86.553
Repeat #0-Fold #9: CPU-86.454 vs Wall-86.465
Although the decision tree does not run in parallel, it can release the Python GIL. This can result in surprising runtime measures as demonstrated below:
with parallel_backend("threading", n_jobs=-1):
run7 = openml.runs.run_model_on_task(
model=dt, task=task, upload_flow=False, avoid_duplicate_runs=False
)
measures = run7.fold_evaluations
print_compare_runtimes(measures)
Repeat #0-Fold #0: CPU-396.746 vs Wall-180.315
Repeat #0-Fold #1: CPU-369.766 vs Wall-160.428
Repeat #0-Fold #2: CPU-341.220 vs Wall-170.745
Repeat #0-Fold #3: CPU-406.798 vs Wall-183.768
Repeat #0-Fold #4: CPU-383.143 vs Wall-187.419
Repeat #0-Fold #5: CPU-341.387 vs Wall-151.239
Repeat #0-Fold #6: CPU-340.346 vs Wall-129.852
Repeat #0-Fold #7: CPU-311.617 vs Wall-110.584
Repeat #0-Fold #8: CPU-163.396 vs Wall-107.557
Repeat #0-Fold #9: CPU-148.019 vs Wall-87.065
Running a Neural Network from scikit-learn that uses scikit-learn independent parallelism using libraries such as MKL, OpenBLAS or BLIS.
mlp = MLPClassifier(max_iter=10)
run8 = openml.runs.run_model_on_task(
model=mlp, task=task, upload_flow=False, avoid_duplicate_runs=False
)
measures = run8.fold_evaluations
print_compare_runtimes(measures)
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sklearn/neural_network/_multilayer_perceptron.py:691: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (10) reached and the optimization hasn't converged yet.
warnings.warn(
Repeat #0-Fold #0: CPU-955.381 vs Wall-955.403
Repeat #0-Fold #1: CPU-1287.133 vs Wall-997.186
Repeat #0-Fold #2: CPU-1246.170 vs Wall-1017.075
Repeat #0-Fold #3: CPU-1288.042 vs Wall-998.478
Repeat #0-Fold #4: CPU-1293.861 vs Wall-1002.083
Repeat #0-Fold #5: CPU-1291.198 vs Wall-999.010
Repeat #0-Fold #6: CPU-1288.308 vs Wall-996.247
Repeat #0-Fold #7: CPU-1294.320 vs Wall-1002.777
Repeat #0-Fold #8: CPU-1289.550 vs Wall-998.240
Repeat #0-Fold #9: CPU-1285.844 vs Wall-995.266
Case 5: Running Scikit-learn models that don’t release GIL¶
Certain Scikit-learn models do not release the Python GIL and are also not executed in parallel via a BLAS library. In such cases, the CPU times and wallclock times are most likely trustworthy. Note however that only very few models such as naive Bayes models are of this kind.
clf = GaussianNB()
with parallel_backend("multiprocessing", n_jobs=-1):
run9 = openml.runs.run_model_on_task(
model=clf, task=task, upload_flow=False, avoid_duplicate_runs=False
)
measures = run9.fold_evaluations
print_compare_runtimes(measures)
Repeat #0-Fold #0: CPU-64.345 vs Wall-64.412
Repeat #0-Fold #1: CPU-64.265 vs Wall-64.441
Repeat #0-Fold #2: CPU-64.076 vs Wall-64.411
Repeat #0-Fold #3: CPU-64.355 vs Wall-66.430
Repeat #0-Fold #4: CPU-61.801 vs Wall-61.920
Repeat #0-Fold #5: CPU-61.847 vs Wall-63.240
Repeat #0-Fold #6: CPU-63.174 vs Wall-63.943
Repeat #0-Fold #7: CPU-64.145 vs Wall-64.287
Repeat #0-Fold #8: CPU-50.552 vs Wall-50.606
Repeat #0-Fold #9: CPU-49.051 vs Wall-49.109
Summmary¶
The scikit-learn extension for OpenML-Python records model runtimes for the CPU-clock and the wall-clock times. The above examples illustrated how these recorded runtimes can be extracted when using a scikit-learn model and under parallel setups too. To summarize, the scikit-learn extension measures the:
CPU-time & wallclock-time for the whole run
A run here corresponds to a call to run_model_on_task or run_flow_on_task
The recorded time is for the model fit for each of the outer-cross validations folds, i.e., the OpenML data splits
Python’s time module is used to compute the runtimes
CPU-time is recorded using the responses of time.process_time()
wallclock-time is recorded using the responses of time.time()
The timings recorded by OpenML per outer-cross validation fold is agnostic to model parallelisation
The wallclock times reported in Case 2 above highlights the speed-up on using n_jobs=-1 in comparison to n_jobs=2, since the timing recorded by OpenML is for the entire fit() procedure, whereas the parallelisation is performed inside fit() by scikit-learn
The CPU-time for models that are run in parallel can be difficult to interpret
CPU-time & wallclock-time for each search per outer fold in an HPO run
Reports the total time for performing search on each of the OpenML data split, subsuming any sort of parallelism that happened as part of the HPO estimator or the underlying base estimator
Also allows extraction of the refit_time that scikit-learn measures using time.time() for retraining the model per outer fold, for the best found configuration
CPU-time & wallclock-time for models that scikit-learn doesn’t parallelize
Models like Decision Trees or naive Bayes don’t parallelize and thus both the wallclock and CPU times are similar in runtime for the OpenML call
However, models implemented in Cython, such as the Decision Trees can release the GIL and still run in parallel if a threading backend is used by joblib.
Scikit-learn Neural Networks can undergo parallelization implicitly owing to thread-level parallelism involved in the linear algebraic operations and thus the wallclock-time and CPU-time can differ.
Because of all the cases mentioned above it is crucial to understand which case is triggered when reporting runtimes for scikit-learn models measured with OpenML-Python!
Total running time of the script: (1 minutes 31.007 seconds)
